Meet Genome Explorer®
Your genomic analysis and clinical decision support platform

EMPOWERING PRECISION MEDICINE
Across our portfolio of products, services, customizable pipelines and reports, Genome International provides clinical decision support tools, revealing crucial information about druggable genes, adverse drug reactions, clinical trials, and more.

OUR WHOLE-GENOME TECHNOLOGIES
Our team has developed many novel technologies to uncover the clinical hotspots in the whole human genome. Unlike traditional software that only focus on the coding regions of genes which comprise less than 2% of the genome, our software targets 100% of the genome, including both coding and non-coding regions. Our methodologies enable scientists to discover the genetic causes of diseases and drug responses throughout the human genome.

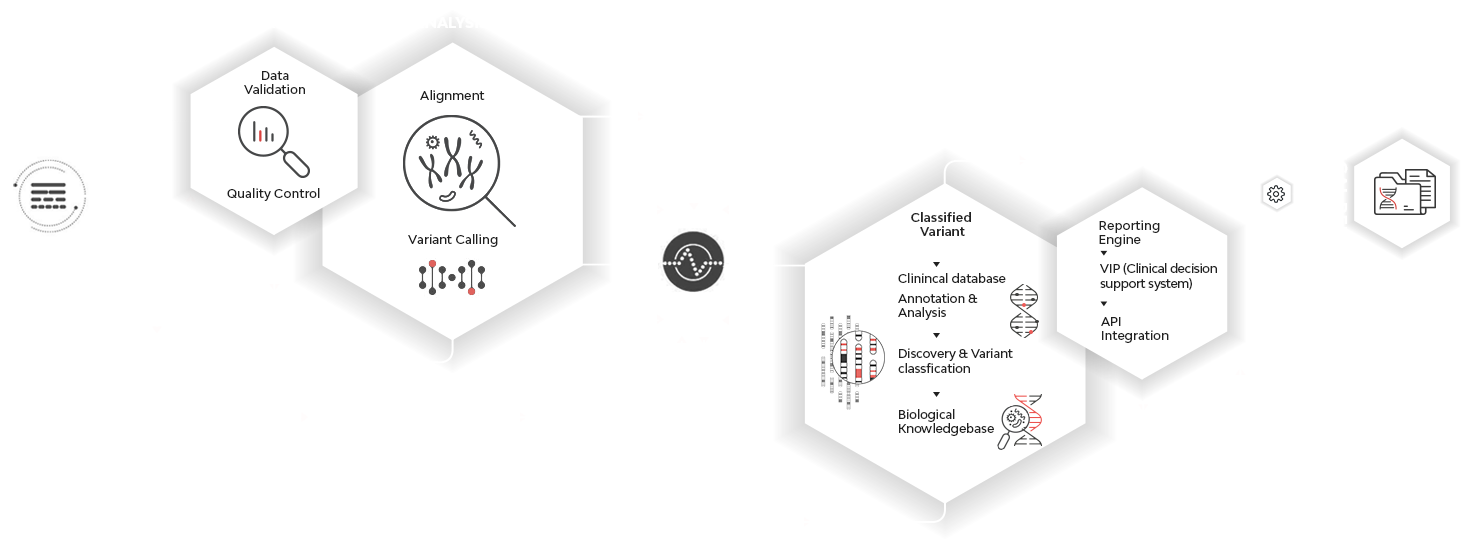
INTELLIGENT WORKFLOW
Customized objective-driven workflows support all NGS experimental strategies with intelligent secondary and tertiary analysis of patient genomes. The clinical interpretations are based on our suite of proprietary algorithms that improve diagnostic accuracy.

SEQUENCING TO DECISION SUPPORT
Genome Explorer performs rapid analysis of the whole genome data and provides an accurate and intuitive clinical report for quick understanding by clinicians. Our platform obtains the patient sequence data from any location, and performs a fast and accurate analysis, enabling clinicians to visualize the results from the patient’s EHR by the touch of a button.

Cancer Diagnosis and Therapeutics

Inherited Disorders