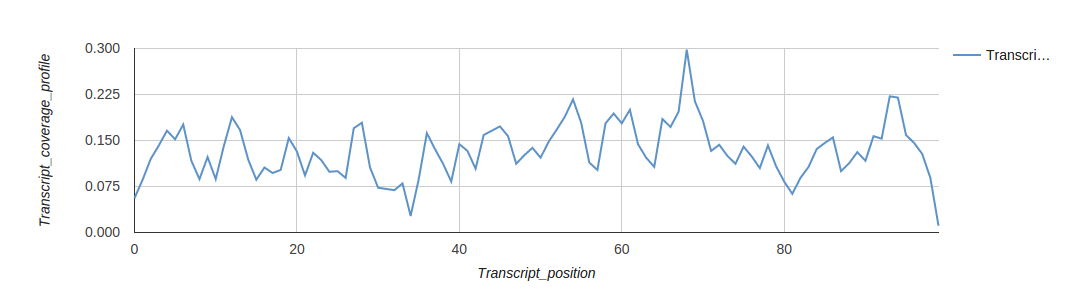
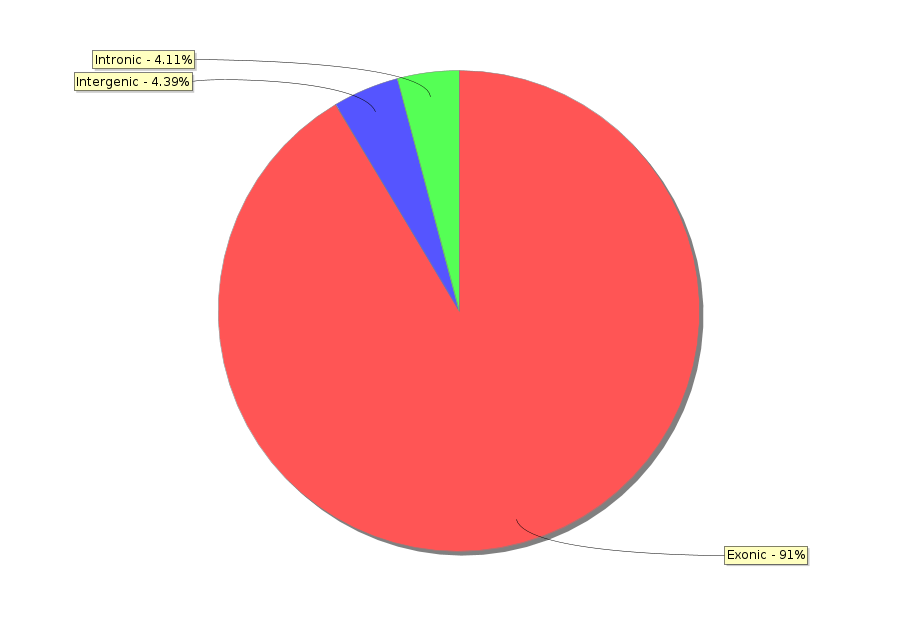
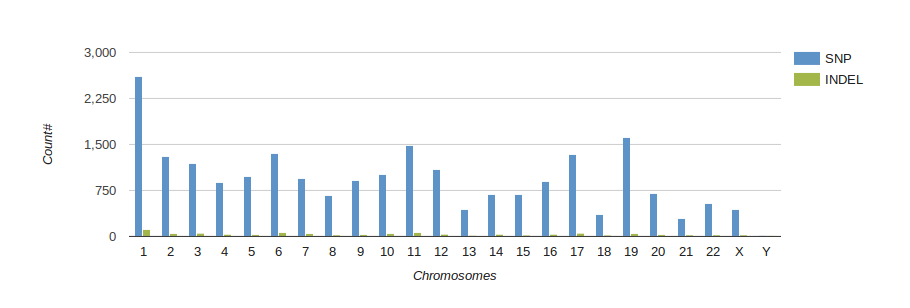
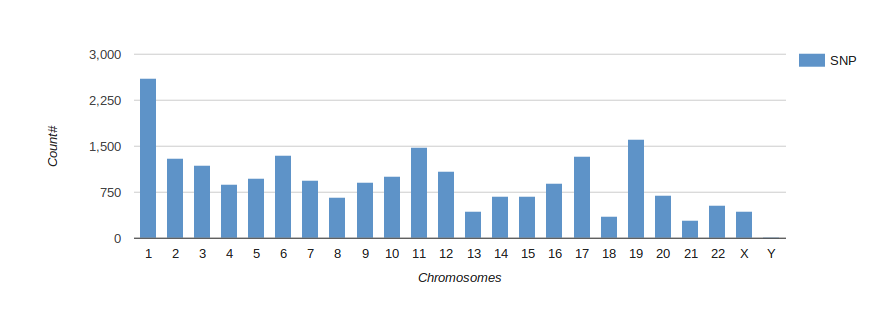
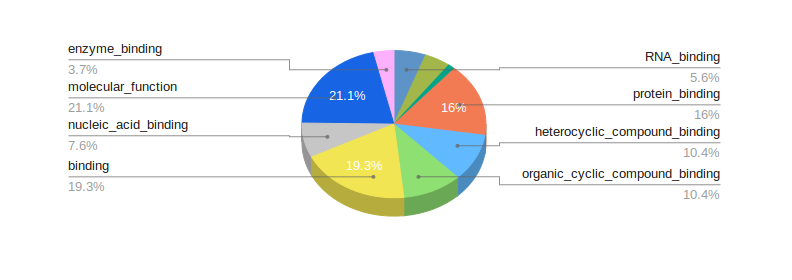
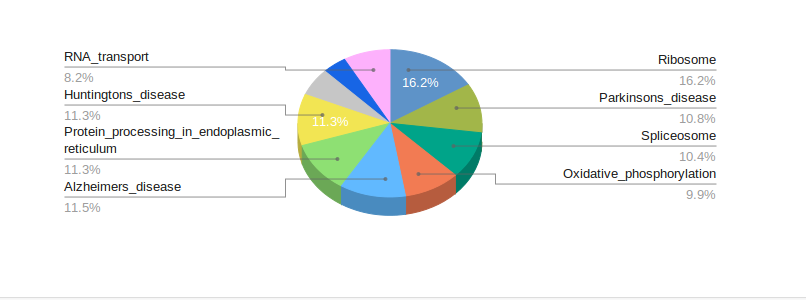
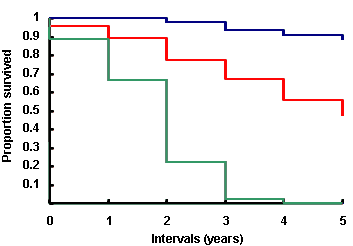
Reports
Turn your complex genomic data into a meaningful report of actionable information and insightful visualizations.
Reports are highly customizable and exportable in industry-class formats.
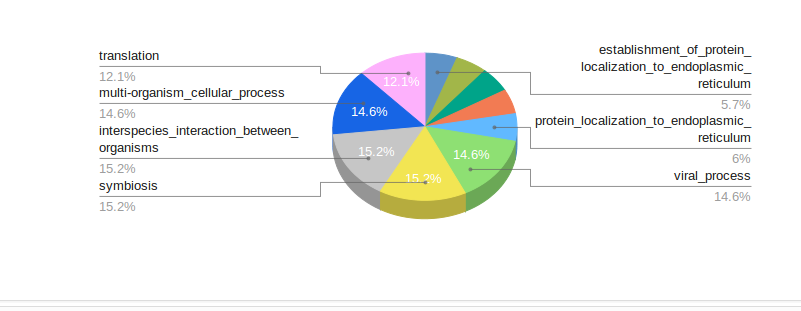
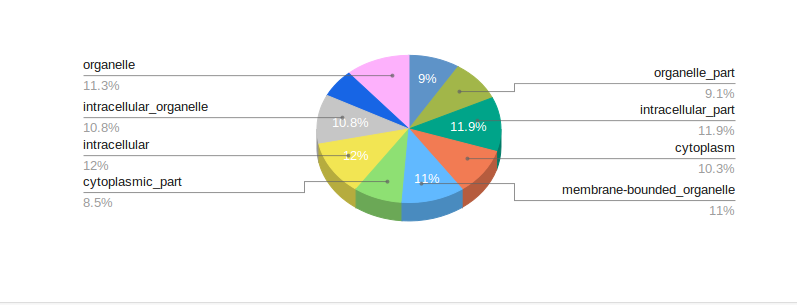
mRNA comprehensive is an extensive workflow to identify differentially expressed genes between your samples, possible gene fusions and transcriptome variants from your RNA-seq data. Downstream analysis such as GO and KEGG-pathway analysis can be performed on the set of interesting genes. In this workflow, parameters can also be optimized depending on datasets and analysis needs. State-of-the-art publication-quality visualization and reports of your analysis results can be easily downloaded in the form of tables, images and html.
Samples are downloaded from amazon s3 bucket.
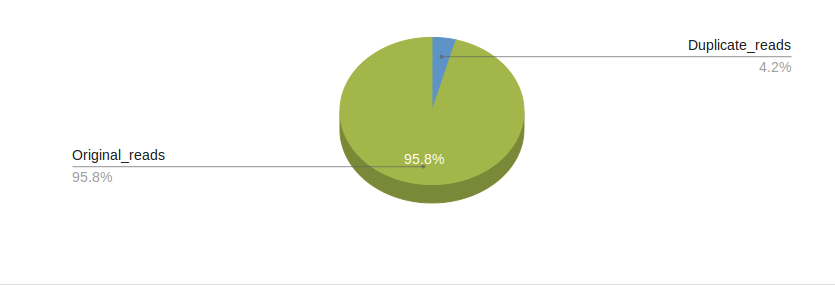
Pre-alignment quality control step is performed to ensure the quality of raw data and it also provides a QC report which can spot biases in the starting library material or during sequencing. We use the FASTQC program which compiles important statistics on NGS data, such as per-base and per-sequence quality scores,GC content and checks for duplicated and overrepresented sequences within the file.
In this step sequencing reads are aligned to the reference genome. The mapping is performed using an ultrafast, memory-efficient read aligner Tophat which is based on Burrows-Wheeler Transformation algorithm. Read alignment information is reported in SAM/BAM format which is used for calculating depth of coverage and calling variants.
Alignment QC step examines the sequencing alignment data in SAM/BAM file format according to the features of the mapped reads and their genomic properties and provides an overall view of the data that helps to detect biases in the sequencing and/or mapping of the data which you should be aware of before doing any further analysis. All statistics about sufficient coverage, depth of sequencing and read counts are provided at this step.
The objective is to assemble reads that were previously aligned to the reference genome into transcript structure (transcript assembly process) and to estimate the abundance levels of genes and transcripts. The expression levels are reported in terms of Fragments Per Kilobase of Exon per Million mapped fragments (FPKM) units.
The expression values of genes and transcripts can be compared across samples post transcript-assembly process.
In this objective, GTF files from each sample are merged together into a single GTF file, which is used as an input to Differential Expression objective.
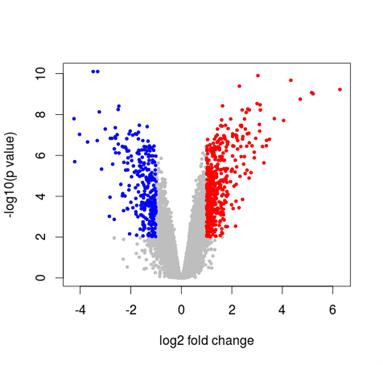
The objective is to find significant changes in gene/transcript expression, splicing, and promoters between two samples in a group.
GATK processing step includes data cleanup operations to correct for technical biases and make the data suitable for analysis. This step involves indel cleaning and base score recalibration following the GATK's latest definition of best practices.
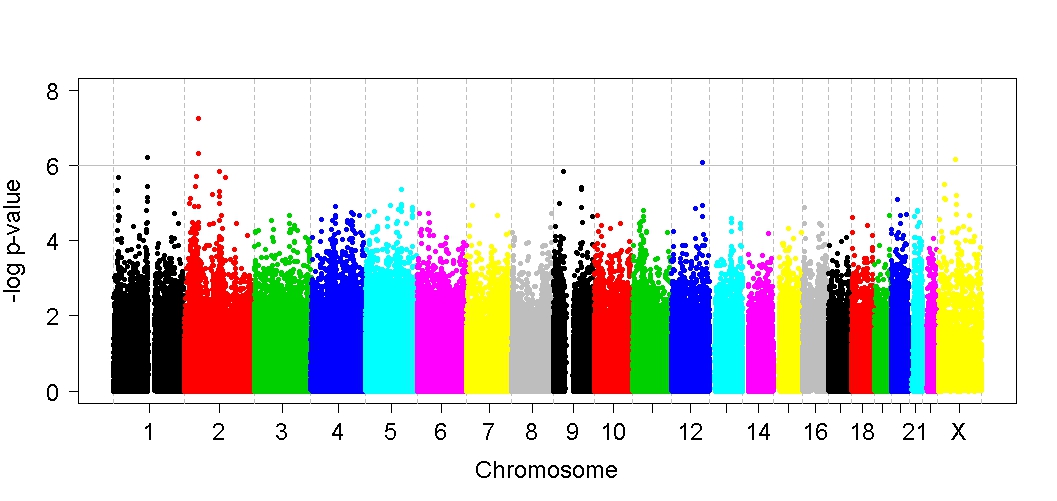
Calling variants using GATK follows the GATK Best Practices to identify and predict the effect of high quality transcriptome variants from your RNA-seq data. Likelihood data for each genotype is calculated and variants are identified with respect to the reference genome. The results are reported in the widely popular VCF format.
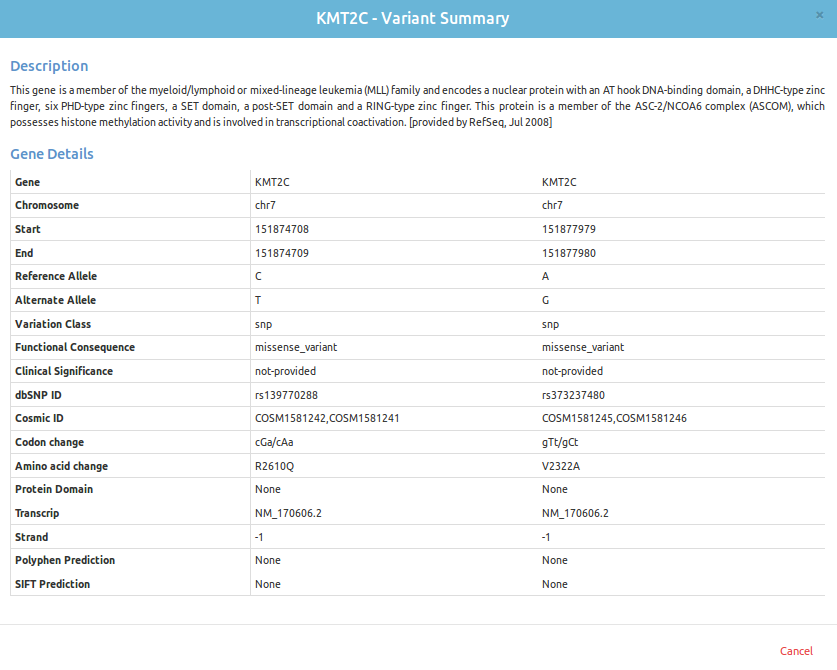
Functional annotation of the high quality variants is performed to predict the effects of variants on genes (such as amino acid changes, etc.) and also it classifies the mutation in various categories like synonoumous or non-synonomous, mis-sense or silent. Novel SNVs are identified by filtering variants that are already annotated in existing publicly available databases like dbSNP. Deleterious variants are separated from the benign ones by assessing the mutagenic potential using SIFT and/or PolyPhen scoring system.
After identification and annotation of high quality single nucleotide variants and small insertions and deletions (indels), these variants are prioritized on the basis of loss of function and actionable mutations. Detailed reports for Novel and Rare loss of function mutations are provided with the added information from databases like omim,clinvar,polyphen and SIFT for clinical significance.
Turn your complex genomic data into a meaningful report of actionable information and insightful visualizations.
Reports are highly customizable and exportable in industry-class formats.
Access and use of the websites or web/mobile applications of Genome International Inc., (“GIC” or “we” or “us”) are governed by the following Terms & Conditions (“T&C”). The T&C document is modified from time to time and the most updated version is made accessible at: http://www.genome.com/terms/. By using websites or web/mobile applications of GIC, you accept that you understand and enter into an agreement or to be bound by this T&C. If you are using our websites or web/mobile applications on behalf of your employer or any other entity and not for individual purposes, by using our websites or web/mobile applications, you also confirm that you are a user authorized to represent your employer or the entity who you represent, enter yourself and your employer as well as the entity that you represent into an agreement or to bind yourself and your employer to this T&C.
GIC websites and web/mobile applications provide and receive certain data ( or “content”) for their individual purposes. All the data on our websites and/or web/mobile applications are proprietary data protected by copyright under both United States and foreign laws and are made available to users in compliance with this T&C only for non-commercial use. Permission to copy, download or store data on our websites or web/mobile applications for your non-commercial use may be granted only with the express written permission of GIC (such as a purchased user subscription, etc.), in compliance with this T&C. Any content of GIC websites and web/mobile applications shall not, in any case, be allowed to be modified, distributed, published, sold, licensed, broadcasted, or used for creating derivative works. By using GIC websites and/or web/mobile applications, you agree to comply with all applicable local, state, federal and international laws, rules and regulations. Users who have written permission from GIC, such as a purchased subscription, are referred to as licensors (or “subscribers”). Our licensors retain all right, title and interest in and to all the data in our websites and web/mobile applications, in compliance with this T&C. Except as expressly stated in this TOU, no rights or licenses are granted in or to the Site or its content (or any other intellectual property of Complete Genomics), by implication, estoppel or otherwise.
GIC does not give warranty or claim the accuracy of data provided on its websites and web/mobile applications, and therefore does not claim any responsibility to update such data. GIC and its associates shall not be held liable for your use of or inability to use its websites and web/mobile applications. GIC and its associates shall not be held liable for any action taken that is based on the data provided on its websites and web/mobile applications. Access to and use of all of GIC’s websites, web/mobile applications and the data they provide is solely at user’s discretion.
GIC websites and web/mobile applications may contain links to third party websites or web/mobile applications in light of their individual purposes. The linked entities are by no means governed by the T&C, rules and regulations of GIC. Therefore, GIC does not endorse, make any warranty or represent their accuracy, legality or timeliness of data provided by them. No part of this T&C and any of GIC policies and procedures shall apply to these third party entities. Accessing and using of these third party websites and web/mobile applications may assume user’s compliance with the terms and conditions or policies and procedures, or rules and regulations applicable to these third party websites and web/mobile applications and is solely at user’s discretion.
GIC respects the privacy of its users and ensures that their personally identifiable information shared with us is given necessary protection and management. In order to address this, GIC has established “Privacy Policy” and users can review and understand it at http://www.genome.com/privacy/. Your use/operation of GIC websites and web/mobile applications is governed by this Privacy Policy. GIC has made detailed information in this Privacy Policy with regard to how we collect, use, disclose, and protect a user’s personal information and other information submitted by a user. You understand that, through your use of GIC websites and/or web/mobile applications, you give consent to the collection, storage and use of such information as described in this T&C and in compliance with our Privacy Policy.
All the content originated from a user, referred to as “user content”, including information, communications, listings, data, text, software, audio files, image files, graphics, video files, messages, etc., is the sole responsibility of the user who shared such content through or by using GIC websites and/or web/mobile applications. A user who shares such content on behalf of one or more entities or persons, including any third party entities, must confirm and warrant that he/she is authorized to act on behalf of and bind such persons or entities to this T&C. As GIC websites and web/mobile applications are used by multiple number of users across globe, GIC is not obliged to any screening, monitoring or controlling the user content posted on this site, including communications, information or other user content originated by users. Such content originating from other users may contain content that is offensive, indecent, inaccurate, objectionable or inappropriate. GIC will not be liable for such user content, including such user information accuracy or errors. GIC will not be liable for any loss or damage incurred as a result of the use of reliance upon content of other users and you agree to release GIC from, and waive any rights to bring or assert any claims for, any liabilities arising from any user content. User is the sole responsible entity for such damages resulting from any infringement of copyright, trademark, or other proprietary right, or any and all harm resulting from your user content or your use of our websites and web/mobile applications in violation of this T&C.
You agree that you are responsible for your own conduct, user content and the consequences thereof while using GIC websites and web/mobile applications. You agree to use our websites and web/mobile applications only for purposes that they are meant for strictly in compliance with the T&C and any applicable laws, rules, regulations, policies and procedures.
By way of example, and not as a limitation, you agree that when using GIC websites and web/mobile applications, you will not offend, abuse, defame, spam, threaten or otherwise violate the legal rights (such as rights of privacy and publicity) of others or harm minors in any way; you will not download any file posted by another that you know, or reasonably should know, cannot be legally distributed in such manner; you will not impersonate another person or entity, or falsify or delete any author attributions, legal or other formal notices, proprietary designations or labels of the origin or source of material; you will not use any information received through GIC websites and web/mobile applications to attempt to identify other users, to contact other users (other than through features for contacting other users that may be offered through them), or for any forensic use; you will not restrict or inhibit any other user from using and enjoying GIC websites and web/mobile applications;
you will not upload, post, provide, email or transmit or otherwise make available
International users agree to comply with their own local rules regarding online conduct and acceptable content, including laws regulating the export of data to the United States or your country of residence.
This Site is not intended for use by minors or designed to attract users under the age of eighteen. GIC will not collect any personally identifiable information from any user under the age of eighteen.
The trademarks, service marks, and logos of GIC that are used and displayed on our websites and web/mobile applications are registered and shall remain the property of GIC. Nothing on GIC websites and web/mobile applications should be construed as granting, by implication, estoppels, or otherwise, any license or right to use any trademark displayed on our websites and web/mobile applications without the prior written permission from GIC. These GIC trademarks may not be used to disparage GIC, its products and services, or otherwise damage any goodwill in GIC trademarks. Use of any GIC trademarks as part of a link to or from any site is prohibited unless GIC permits such action in a written communication.
GIC respects and promotes its user to respect the intellectual property rights of others. We will respond to notices of alleged copyright infringement that comply with applicable law and are properly provided to us. If you believe that any work of authorship that you own or control has been copied or utilized on our websites and web/mobile applications in a way that constitutes copyright infringement, please provide us with the following information in accordance with the Digital Millennium Copyright Act: (i) a physical or electronic signature of the copyright owner or a person authorized to act on their behalf; (ii) identification of the copyrighted work claimed to have been infringed; (iii) identification of the material that is claimed to be infringing or to be the subject of infringing activity and that is to be removed or access to which is to be disabled, and information reasonably sufficient to permit us to locate the material; (iv) your contact information, including your address, telephone number, and an email address; (v) a statement by you that you have a good faith belief that use of the material in the manner complained of is not authorized by the copyright owner, its agent, or the law; and (vi) a statement that the information in the notification is accurate, and, under penalty of perjury, that you are authorized to act on behalf of the copyright owner.
Notice of alleged copyright infringement or other legal notices regarding user content appearing on our websites and web/mobile applications shall be sent to [support@gna.genomels.com]. We reserve the right to remove user content alleged to be infringing or otherwise illegal without prior notice.
GIC updates, revises or amends this T&C in its sole discretion and notifies its users at least fifteen (15) days in advance before bringing the new T&C into effect, by posting the modified or amended T&C on our websites and web/mobile applications. After the given fifteen (15) days period, or by its sole discretion, GIC will bring all modifications in this T&C into effect. In cases where user finds any or all of the revised T&C, user will have to stop using GIC websites and web/mobile applications. By continuing to access or use them after the T&C are revised, you agree to be bound by the revised T&C.
If you use this site, you are responsible for maintaining the confidentiality of your account and password and for restricting access to your computer, and you agree to accept responsibility for all activities that occur under your account or password. GIC and its associates reserve the right to refuse service, terminate accounts, remove or edit content, or cancel orders at their sole discretion.
GIC WEBSITES AND WEB/MOBILE APPLICATIONS ARE PROVIDED ON AN "AS IS" AND GIC MAKES NO REPRESENTATIONS OR WARRANTIES OF ANY KIND, EXPRESS OR IMPLIED, AS TO THE OPERATION OF THIS SITE OR THE INFORMATION, CONTENT, MATERIALS, OR PRODUCTS INCLUDED ON THIS SITE. YOU EXPRESSLY AGREE THAT YOUR USE OF GIC WEBSITES AND WEB/MOBILE APPLICATIONS IS AT YOUR SOLE RISK. TO THE FULL EXTENT PERMISSIBLE BY APPLICABLE LAW, GIC DISCLAIMS ALL WARRANTIES, EXPRESS OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. GIC DOES NOT WARRANT THAT ITS WEBSITES, WEB/MOBILE APPLICATIONS, SERVERS, OR E-MAIL SENT FROM GIC ARE FREE OF VIRUSES OR OTHER HARMFUL COMPONENTS. GIC WILL NOT BE LIABLE FOR ANY DAMAGES OF ANY KIND ARISING FROM THE USE OF ITS WEBSITES AND WEB/MOBILE APPLICATIONS, INCLUDING, BUT NOT LIMITED TO DIRECT, INDIRECT, INCIDENTAL, PUNITIVE, AND CONSEQUENTIAL DAMAGES.
Any dispute relating in any way to your GIC websites and web/mobile applications you purchase through GIC shall be submitted to confidential arbitration in Delaware, USA, except that, to the extent you have in any manner violated or threatened to violate GIC's intellectual property rights, GIC may seek injunctive or other appropriate relief in any state or federal court in the state of Wisconsin, USA, and you consent to exclusive jurisdiction and venue in such courts. Arbitration under this agreement shall be conducted under the rules then prevailing of the American Arbitration Association. The arbitrators award shall be binding and may be entered as a judgment in any court of competent jurisdiction. To the fullest extent permitted by applicable law, no arbitration under this Agreement shall be joined to an arbitration involving any other party subject to this Agreement, whether through class arbitration proceedings or otherwise.
Please review our other policies and disclaimers. These policies also govern your visit to GIC websites and web/mobile applications. We reserve the right to make changes to our websites, web/mobile applications, policies, and these T&C at any time. If any of these conditions shall be deemed invalid, void, or for any reason unenforceable, that condition shall be deemed severable and shall not affect the validity and enforceability of any remaining condition.
GIC requests and encourages its users to report any violations of this T&C and our other policies or provide any comments or questions by emailing us at [support@gna.genomels.com]. You agree, however, that If you make or provide any suggested improvements, enhancements, comments, recommendations or other feedback with respect to GIC websites, web/mobile applications or its other products and services , you hereby grant to GIC an unlimited, royalty-free, worldwide, irrevocable, perpetual, fully sublicensable, transferable license to use and exploit such suggestions in any way for any purpose under any intellectual property rights in, to and under such suggestions you may have.
GIC, Inc. knows that you care how we use and protect the data you submit to us and other information we may gather from you. To show our commitment to your privacy and to explain how we use and protect your information, GIC has implemented this privacy policy ("Privacy Policy").
Your use of our services and the GIC website (the "Site") is voluntary; if you do not agree with any of the terms and conditions of this Privacy Policy, please do not submit your data on any of our websites (collectively the “Sites”). This privacy policy applies only to the data/information obtained by our sites from the user, but not data/information obtained by any other means (online or offline).
When you inquire about our offerings, register for our services or request access to our web applications on our Site and during your use of the Site and services, we may collect information that identifies you as an individual ("personally identifiable information"), such as: your name, company or organization name, title, email address, postal address and telephone numbers. In case of purchasing product subscriptions or any services, we may ask for billing information, such as credit card details. If you contact us, we may keep a record of all or some of this data for reference in case of your follow-up communication with us.
For maintenance, updating and administration of our websites, web applications and other related services, our servers may automatically collect your usage data. This data includes the browser you use, operating system on your your computer, your Internet service provider’s domain details, your Internet Protocol (IP) address, pages you visited on our Site and other Site usage patterns.
Cookies are used to track your activity on our sites and they enable us to give you the best and enhanced website functionality experience. Cookies are small data chunks that our servers send to your browser while using our sites, and are saved on the hard drive of your computer. Most browsers come with settings that enable you to accept/reject cookies from any site. Rejecting all cookies, however, may influence your experience on our sites. If you choose to accept cookies from us and our associates, you are agreeing to let us and our associates install cookies on your computer.
GIC collects and stores the genomic sequence data that you submit to the Site to implement any or all of analysis pipelines, which may have been derived from a sample taken from a human or other organism, as well as metadata and other information related to the sequence data that you provide in connection with your use of our services. We rely on you to obtain all necessary permissions and informed consents from the donors of samples from which you derive the sequence data you submit. We also rely on you to insure that all sequence data derived from human samples have been de-identified by removing any information that would enable GIC or a third party to associate the sequence data with a specified individual.
We take organizational, technical and administrative measures designed to protect the information you provide through the Site that is under our control. For example, only our employees who need the information to perform a specific job are granted access to personally identifiable information and all employees are kept up-to-date on our security and privacy practices. Further, we use Secure Socket Layer (SSL) 2048 bit encryption on all web pages where your personally identifiable information is collected or where you upload sequence data to protect its transmission over the Internet. Unfortunately, however, no data transmission over the Internet or data storage system can be guaranteed to be 100% secure, so as a result, while we strive to protect your information, we cannot ensure the security of information you transmit to us, we transmit to you or that we store. If at any time you believe that your interaction with the Site is no longer secure, please let us know as soon as possible. Further, if you register and create a profile on the Site, your profile and the data submitted by you to the Site are password-protected. We recommend that you do not divulge your password to anyone. Also remember to sign out of your GIC account and close your browser window when you have finished your work. This is to prevent others from accessing your information and data.
We may use personally identifiable information about you to contact you and provide services to you, at your request. We may use it to provide you with a personalized experience on our Site and to communicate with you through periodic messages regarding new service features and related products and services provided by our partners, events, and other information and notices we believe you may find interesting or useful. Your credit card information (if applicable) may be used by our financial services providers only for payment processing and fraud prevention.
We use the automatic usage information collected to maintain, secure and improve our Site, and to understand your interests when visiting our Site.
We may generate statistical information regarding our user-base and use it to analyze our business. We also may use users' raw sequence data in the aggregate to improve our algorithms. We might share such statistical or aggregate information with third parties, but it doesn't include any personally identifiable information or individual sequence sample.
We do not use the genomic information you provide to us or any reports generated by your analysis of that information using tools available on the Site for any purpose other than providing services to you.
We store the sequence data you submit and your and account information (including personally identifiable information) on "cloud" servers owned and operated by a third party provider. The current provider is Amazon Web Services (AWS). For information about AWS's privacy protection and data security practices, please visit http://aws.amazon.com/security. We also may rely on various service providers, business partners and other contractors to provide services that support the Site and our operations, including, without limitation, maintenance of our databases, distribution of emails and newsletters on our behalf, data analysis, payment processing and other services of an administrative nature. Such third-parties may have access to your personally identifiable information for the purpose of performing the service for which they were engaged. These third parties are committed to protecting the privacy of our users, will implement reasonable safeguards to protect your personally identifiable information and are required to utilize your information only for the purposes for which it was provided. GIC is not responsible or liable for the use of personally identifiable information by such third- parties.
We may disclose personally identifiable information and other user information in special cases when we have reasons to believe that disclosing this information is necessary to identify, contact or bring legal action against someone who may (either intentionally or unintentionally) be causing injury to or interference with our rights or property, users of our Site, or anyone else who could be harmed by such activities. We may also disclose user information when we believe in good faith that such disclosure is required by applicable law.
Lastly, we reserve the right to transfer any and all information that we collect from Site users to a third-party in the event of any corporate reorganization, merger, sale, joint venture, assignment, transfer or other disposition of all or any portion of GIC' business, assets or stock.
Our Site allows you to share the sequence data and other information you submit to the Site with other users of the Site. GIC is not responsible for such other users' use of your sequence data and other information. You understand that information that you share might be copied or re-shared by other users. Even after you remove information from your account or delete your account, copies of that information may remain viewable elsewhere to the extent it has previously been shared with others.
We and our service providers may make available through the Site certain services to which you are able to post information and materials (for example, forums, chat rooms, message boards, and news groups). If you visit such a service on a Site, please remember that any information that is disclosed in these areas becomes public information and may be available to users of the Site and to the general public. In addition, when you choose to make a posting on such services, certain personally identifiable information (such as your username) will be available for other users to view.
We urge you to exercise discretion and caution when deciding to disclose personally identifiable information, or any other information, on the Site. We are not responsible for the use of any personally identifiable information or other information you voluntarily disclose or make available to other users or the public through the site.
If you no longer wish to receive new product/service notices, email newsletters or other future communications from us, please follow the opt-out instructions included in each communication or notify us by email at support@genome.com with the word "remove" in the subject header, and we will remove your name from our email recipient list. You may not, however, opt out of necessary service or account maintenance notices or other administrative and transactional notices.
This Site and our services are not directed towards users under the age of 13 and we do not knowingly collect personally identifiable information online from users under the age of 13.
On the Site, we may provide links to websites maintained by third-parties, which we believe you may find useful. GIC is not responsible for the privacy practices of these other websites and we encourage you to review the privacy policies of each of those other websites. We make no representation with regard to the policies or business practices of any websites to which you connect through a link from this Site, and are not responsible for any material contained on, or any transactions that occur between you and, any such website.
We host and maintain this Site in the United States. By providing personally identifiable information and other information to this Site, you understand and consent to the collection, use, processing and transfer of such information to the United States and other countries or territories, which may not offer the same level of data protection as the country where you reside, in accordance with the terms of this Privacy Policy.
Under California Civil Code Section 1789.3, California Site users are entitled to the following specific consumer rights notice: The Complaint Assistance Unit of the Division of Consumer Services of the California Department of Consumer Affairs may be contacted in writing at 400 R Street, Suite 1080, Sacramento, California 95814, or by telephone at (916) 445-1254 or (800) 952-5210.
GIC is not a health plan, health care provider, or health care clearinghouse as defined in the Health Insurance Portability and Accountability Act of 1996 and its related regulations set forth in Parts 160, 162, and 164 of Title 45 of the Code of Federal Regulations (collectively, "HIPAA") and GIC is not acting on behalf of a one of those entities as the operator of the Site. In addition, GIC does not collect "individually identifiable health information" as defined by HIPAA. As a consequence, neither the genomic information you submit nor any other information we collect will be subject to the limitations on the use and disclosure of such information provided by HIPAA.
Please note that we review our privacy policies from time to time and as such we may revise this Privacy Policy, at our sole discretion, by posting an amended policy on the Site. We ask that you periodically review this page to ensure continuing familiarity with the most current version of our Privacy Policy. Any changes to our Privacy Policy will become effective upon our posting of the revised Privacy Policy on the Site. Use of the Site following such changes constitutes your acceptance of the revised Privacy Policy then in effect. You will be able to determine when this Privacy Policy was last revised by checking the "Last updated" information that appears at the top of this page.
We are interested in answering your questions or hearing any concerns regarding our Privacy Policy and data protection practices. If you wish to contact us, please email us at support@genome.com.
 in the last column of samples table and click on it.
in the last column of samples table and click on it.